Every year, millions of patients suffer from diseases that lack effective treatments. Despite over 4,000 FDA-approved drugs on the market, the vast majority of the estimated 18,500 diseases remain without approved therapies. Traditional drug development is costly and time-consuming, but what if the answers to many of these unmet medical needs already exist within our current arsenal of medicines?
At Every Cure, we are pioneering a groundbreaking field known as computational pharmacophenomics—a systematic, data-driven approach to uncovering new uses for existing drugs. This transformative methodology builds on years of drug repurposing research, integrating massive biomedical datasets with advanced machine learning to predict which medications might be effective for diseases beyond their original indications.
The Power of Drug Repurposing
Drug repurposing—the practice of using existing medicines to treat different diseases—has already changed the landscape of medicine. Drugs like tocilizumab, originally developed for idiopathic multicentric Castleman disease (iMCD), have since been repurposed for rheumatoid arthritis, cytokine release syndrome, and even severe COVID-19. This is because many diseases share underlying biological mechanisms that can be targeted with the same drugs.
However, despite the promise of repurposing, the process has historically been sporadic and reliant on chance discoveries. Physicians legally prescribe drugs off-label, but there has been no centralized, systematic effort to identify and validate the best repurposing opportunities—until now.
A Systematic Approach: Computational Pharmacophenomics
Every Cure is building a comprehensive, AI-powered drug repurposing platform that systematically evaluates the potential of all drugs across all diseases. Our approach is unique because it is both drug-agnostic and disease-agnostic, meaning it does not start with a single disease or a single drug but instead evaluates all possible drug-disease combinations.
Using knowledge graphs and machine learning algorithms, our platform analyzes connections between drugs, diseases, genes, and molecular pathways. One of our core tools, KGML-xDTD, employs AI models to rank the likelihood of a drug treating a specific disease, allowing us to prioritize the most promising candidates for further study. This approach has already led to life-saving discoveries—including treatments that have rescued critically ill patients who had exhausted all other options.
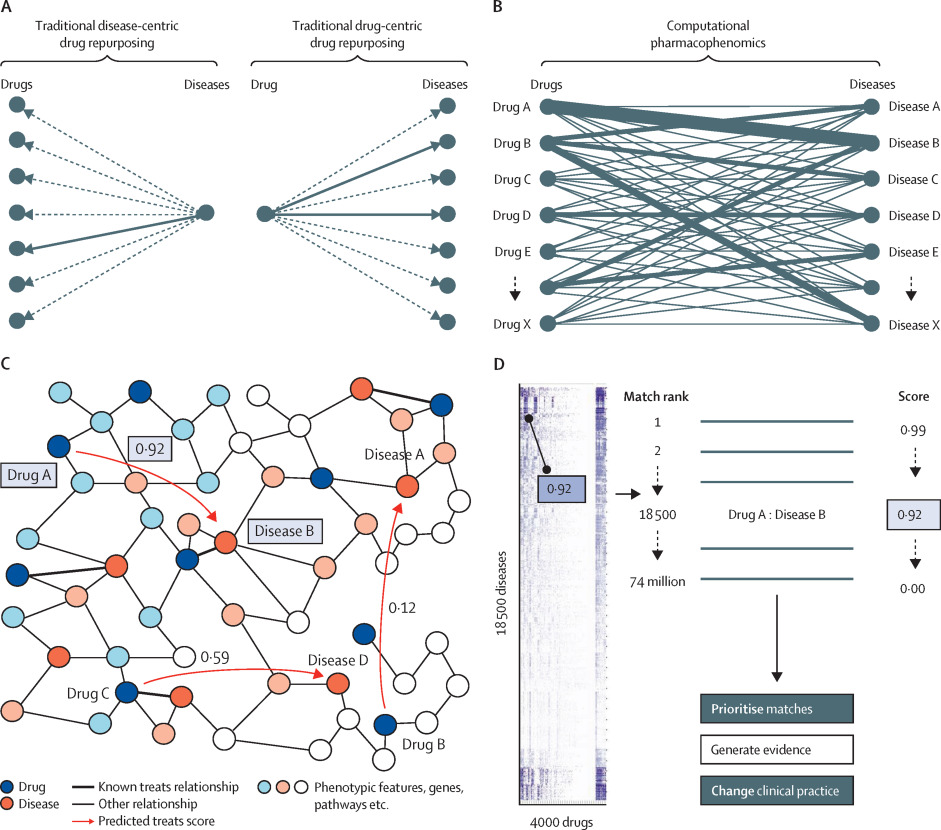
Figure: Computational pharmacophenomics quantifies the likelihood of every approved drug to be able to treat every disease
(A) Visualisation of traditional drug repurposing (single drug or single disease approach). (B) Visualisation of computational pharmacophenomics (systematic quantitative evaluation of the potential of all drugs against all diseases). (C) A sample portion of a knowledge graph representing biomedical concepts (nodes) and the relationships between them (edges). Machine learning models are trained on known treats relationships (thick lines) and applied to the rest of the graph to quantify the likelihood that the new drug–disease match would be an effective treatment (red arrows). (D) Heatmap of every drug–disease match, which can be exported into a match rank list to prioritise matches for further research
A Global Drug Repurposing Database
By 2026, Every Cure will publicly release putative efficacy scores for every drug-disease match, creating an unprecedented open-access resource for physicians, researchers, and policymakers. This tool will:
- Identify high-impact repurposing opportunities for clinical trials
- Highlight underutilized treatments that already have strong clinical evidence but lack awareness
- Provide physicians with evidence-based insights to guide off-label prescribing
From Predictions to Patient Impact
A key differentiator of Every Cure’s platform is its real-world validation process. Unlike traditional drug development, which can take decades, we leverage existing clinical data and real-world evidence to accelerate the transition from AI-driven predictions to clinical adoption.
For example, when a patient with POEMS syndrome was preparing for hospice care due to a lack of treatment options, our platform identified multiple high-scoring multiple myeloma drugs that could be repurposed. These treatments saved the patient’s life—demonstrating the real-world power of computational pharmacophenomics.
A Call to Action
Every Cure is committed to eliminating the treatment gap by ensuring that every possible use of an existing drug is explored. But we can’t do it alone. We invite researchers, clinicians, and patients to contribute insights and collaborate in our mission to make life-saving treatments accessible to all.
Visit EveryCure.org/experts to learn more, explore our research, and join the movement to revolutionize drug repurposing. Together, we can unlock the full potential of medicine and transform the future of healthcare.
Read full paper: https://www.thelancet.com/journals/lanhae/article/PIIS2352-3026(24)00278-3/fulltext
